Data processing and analysis
Data processing and analysis#
Issue
Researchers typically execute a set of signal pre-processing steps prior to advanced data analysis, to, for instance, identify and remove noise, align data spatially and temporally, segment spatio-temporal regions of interest, identify patterns and latent signal structures (e.g., clustering), integrate the information from several modalities, introduce prior knowledge about the device or the physiology of the specimen, etc. The combination of the operations that take the unprocessed data as the input, prepare the data for analysis, and finally, perform advanced analysis, comprise a full analysis pipeline or workflow. In implementing such analysis workflows, software has emerged as a critical research instrument greatly relevant to ensure the reproducibility of studies.
What do we provide
TBE.
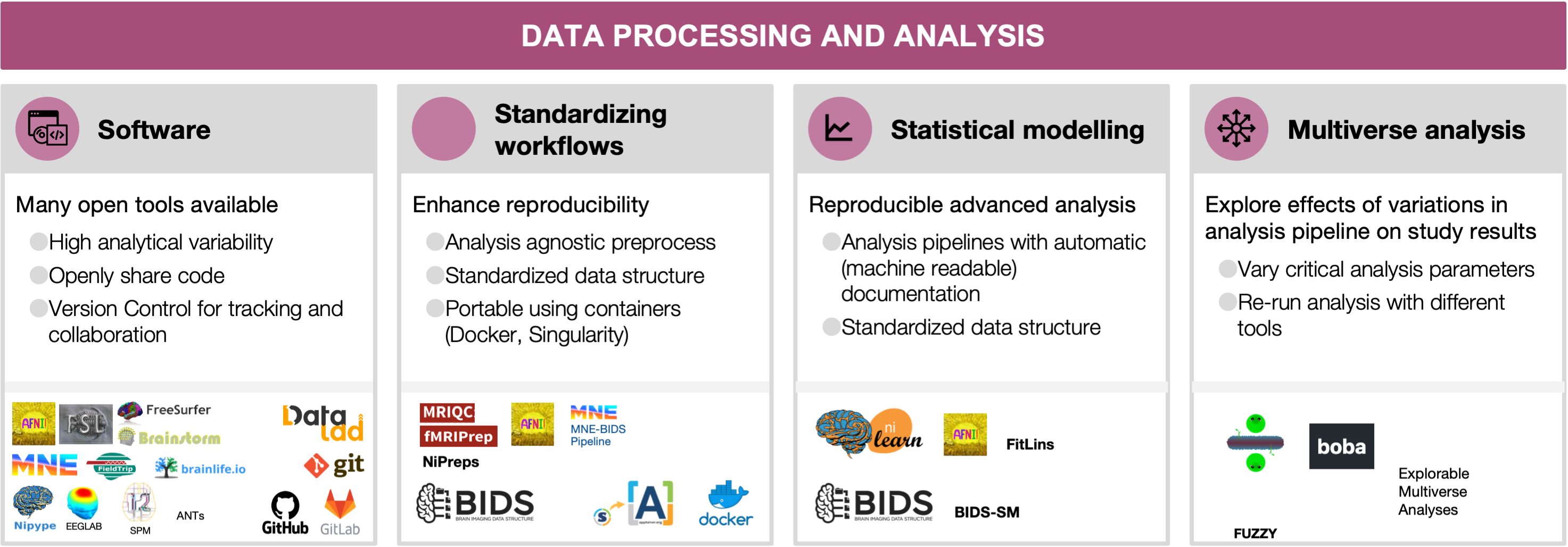
5.1 Software as a research instrument
The digital nature of neuroimaging data along with the large, and constantly increasing, net amounts of daily acquired data, place software as a central instrument of the neuroimaging research workflow. As a result, many toolboxes containing utilities ranging from early steps of preprocessing to statistical analysis and visualization of results have emerged, and some have largely shaped the software development in the field, e.g., AFNI [Cox, 1996, Cox and Hyde, 1997], FSL [Jenkinson et al., 2012], SPM [Flandin and Friston, 2008, Litvak et al., 2011, Penny et al., 2011], FreeSurfer [Dale et al., 1999, Dale and Sereno, 1993], Brainstorm [Tadel et al., 2011, Tadel et al., 2019], EEGLAB [Delorme and Makeig, 2004, Delorme et al., 2021], MNE-Python [Gramfort et al., 2014, Gramfort et al., 2013], FieldTrip [Oostenveld et al., 2011] (see the resources table). More recently, some software packages have been developed to cover additional aspects of the neuroimaging workflow. For instance, nibabel [Brett et al., 2020] to read and write images in many formats, the Advanced Normalization Tools (ANTs) for image registration and segmentation, or Nilearn [Abraham et al., 2014] for statistical analysis and visualization. Workflow engines conveniently connect between the building blocks and determine how the steps are executed in the computational environment. Solutions range from general-purpose scripting (e.g., Bash or Python) to neuroimaging-specific libraries (e.g., NiPype; [Gorgolewski et al., 2011]). Researchers have all these tools (and others) at their disposal to “mix-and-match” in their workflow. Therefore, ensuring the proper development and operation of the software engine is critical to ensure the reproducibility of results [Tustison et al., 2013]).
Relatedly, the variety of software implementations is an additional motive of concern. As remarked by Carp ([Carp, 2012, Carp, 2012]) based on the analysis of thousands of fMRI pipelines, analytical flexibility in combination with incomplete reporting precludes the reproducibility of the results. A recent comprehensive investigation, the Neuroimaging Analysis Replication and Prediction Study (NARPS; [Botvinik-Nezer et al., 2020]) found that when 70 different teams were asked to analyze the same fMRI data to test the same hypotheses, each team chose a distinct pipeline and results were highly variable. Other studies suggest similar problems in EEG [Clayson et al., 2021, Šoškić et al., 2021], PET [Nørgaard et al., 2020] and diffusion MRI [Schilling et al., 2021].
There are two crucial aspects of the high analytical variability and its effect on results in neuroimaging. First, when high analytical variability (that potentially affects results) is combined with partial reporting or with incentives to find significant effects, it can alarmingly undermine the reliability and reproducibility of results. Second, even in the apparently ideal scenario in which the researcher performs a single pre-registered valid analysis and reports it fully and transparently, it is still likely that the results are not robust to arbitrary analytical choices. Therefore, new tools are needed to allow researchers to perform a “multiverse analysis” (section 5.4), where multiple data workflows are used on the same dataset and all the results are reported and their agreement or convergence discussed. Community-led efforts to develop high-quality “gold standard” data workflows may also reduce researchers’ degrees of freedom as well as accelerate data analysis, although different pipelines may be optimal for different research questions and data.
Nevertheless, neuroimaging researchers frequently encounter gaps that readily available toolboxes do not cover. These gaps, amongst a number of other reasons (e.g., deploying a data workflow on a high-performance computer), pushes researchers into creating their own software implementations. However, most neuroimaging researchers are not formally trained in related fields of computer science, data science, or software engineering, and formal software development practices are often not included in undergraduate or graduate level neuroimaging training. This mismatch often results in undocumented, hard to maintain, and disorganized code; largely as a consequence of unawareness of software development practices. It also increases the likelihood of undetected errors that may remain even after running tests on the code.
The first and foremost strategy available to maximize the transparency of research methods is openly sharing the code with the minimal restrictions possible (see Section 6.2; [Gorgolewski and Poldrack, 2016, Barnes, 2010]). Complementarily, version control systems, such as Git (Blischak, Davenport, and Wilson 2016, see the resources table), are the most basic and effective tool to track how software is developed, and to collaboratively produce code. Beyond making the code available to others, software tools can implement further transparency strategies by thoroughly documenting their tools and by supporting implementations with scientific publications ([Gorgolewski and Poldrack, 2016, Barnes, 2010]).
5.2 Standardizing preprocessing and workflows
Although the diversity in methodological alternatives has been key to extracting scientific insights from neuroimaging data, appropriately combining heterogeneous tools into complete workflows requires substantial expertise. Traditionally, researchers used default workflows distributed along with individual software packages, or alternatively, individual laboratories have developed in-house analysis workflows that resulted in highly specialized pipelines. Such pipelines are often not thoroughly validated and difficult to reuse due to lack of documentation or accessibility to outside labs. In response, several community-led efforts have spearheaded the development of robust, standardized workflows.
An early effort towards workflow standardization was the Configurable Pipeline for the Analysis of Connectomes (C-PAC; [Craddock et al., 2013]), which is a “nose-to-tail” preprocessing and analysis pipeline for resting state fMRI. C-PAC offers a comprehensive configuration file, editable directly with a text editor or through C-PAC’s graphical user-interface, prescribing all the tools and parameters to be executed, and thereby making strides towards keeping methodological decisions closely traced. Similarly, large-scale acquisition initiatives released workflows tailored for their official imaging protocols (e.g., the HCP Pipelines [Glasser et al., 2013] and the UK Biobank [Alfaro-Almagro et al., 2016]).
Conversely, fMRIPrep [Esteban et al., 2019] proposed the alternative approach of restricting the pipeline goals to the preprocessing step, while accepting the maximum diversity possible of the input data (i.e., not tailored to a particular experimental design or analysis-agnosticity). This approach has recently been proposed for additional modalities (e.g., dMRI, ASL, PET) and population/species of interest (e.g., fMRIPrep-rodents, fMRIPrep-infants) under a common framework called NiPreps (NeuroImaging PREProcessing toolS). NiPreps is a community-led endeavor with the goal of ensuring the generalization of the building blocks of preprocessing across modalities (e.g., the alignment of fMRI and dMRI with the same participant / animal’s anatomical image) and specimens (e.g., using the same brain extraction from anatomical data using the same algorithm and implementation on both human adults and rodents). Similar standardization efforts are starting to be adopted for EEG [Desjardins et al., 2021] and MEG (e.g., MNE-BIDS pipeline; [Jas et al., 2018]). Further examples of standardized workflows are found in the resources table.
An additional and relevant premise of standardized workflows is transparency — tools must be transparent not only in their implementation, but also in their reporting. For example, fMRIPrep produces visual reports with the double goal of assessing the quality of results, and also providing the researcher with a resource to comprehensively understand every step of the workflow. In addition, the report includes a text description which comprehensively describes each major step in the pipeline, including the exact software version and principle citation. This text, referred to as the “citation boilerplate”, is released under a public domain license, and therefore can be included verbatim in researcher’s manuscripts, facilitating accurate reporting and proper referencing of academic software. A final relevant aspect towards transparency is the comprehensive documentation of pipelines.
In most cases, standardized workflows preprocess datasets in a fully automated manner, taking a BIDS dataset as input and outputting data that is ready for subsequent analysis with little manual intervention. Importantly, such workflows are typically designed to be as robust as possible to diverse input data (e.g., with varying parameters or sampling distant populations), a challenge that is facilitated by data standardization (i.e., BIDS). Additionally, workflows must be portable, enabling users to execute them in a wide variety of environments. A key technology in this endeavor is containers—such as Docker and Apptainer/Singularity—which facilitate packaging specific versions of heterogeneous dependencies while ensuring cross-platform compatibility (e.g., high-performance computing clusters, desktop, or cloud services). The BIDS apps framework (Section 4.1) leverages containers by standardizing input parameters to make it trivially easy to execute a wide variety of standardized workflows on BIDS datasets. An example of a higher-level combination of workflows is found in Esteban et al. Esteban et al. [2020], which describes an MRI research protocol using MRIQC and fMRIPrep. Finally, recent efforts to standardize the outputs of workflows (BIDS Derivatives), further enhances the interoperability of workflows, by ensuring their outputs are compatible with subsequent analysis.
5.3 Statistical modeling and advanced analysis
Analysis of neuroimaging data is particularly heterogeneous and prone to excessive analytical flexibility and underspecified reporting [Carp, 2012, Carp, 2012]. Whereas preprocessing is ideally performed once per dataset, there is often a large number of types of analyses that may be used with the preprocessed data. In MRI and fNIRS, for example, analyses range from multi-stage general linear models (GLMs), multivariable decoding analyses, to anatomical and functional connectivity, and more. In PET, analyses consist of region-wise averaging, although voxel-wise approaches are gaining popularity, followed by kinetic modeling and subsequent statistical analyses, which can be GLM or more advanced, such as latent variable models. In MEG and EEG, the broad variety includes analyses such as evoked response potentials, power spectral density, source reconstructions, time-frequency, connectivity, advanced statistics and more. Each type of analysis also has a wide variety of subtypes, parameters, and statistical models that can be specified, and the form of that specification varies across the dozens of analysis packages that implement each type of analysis.
Data analysis reporting may be made more transparent by sharing code that relies on open-source software. A prime example is SPM [Flandin and Friston, 2008], which has been open source since its inception in 1991. Additional widely used open-source tools for data analysis are FSL and AFNI for MRI, and some examples of reproducible pipelines for MEG and EEG developed based on each of the following software: EEGLAB [Pernet et al., 2020], Fieldtrip [Andersen, 2018, Meyer et al., 2021, Popov et al., 2018], Brainstorm [Niso et al., 2019, Tadel et al., 2019], SPM [Henson et al., 2019] and MNE-Python [Andersen, 2018, Jas et al., 2018, van Vliet et al., 2018] (see [Niso et al., 2022] for a detailed review on main EEG and MEG open toolboxes and reproducible pipelines). Reproducibility is also improved when relying on modular and well-documented software such as Nilearn, which offers versatile methods to perform advanced analyses of fMRI data, from GLM to connectomic and machine learning [Abraham et al., 2014]. Ideally, a single analysis script is made creating a report, from signal extraction, data analysis, and reproducing all figures.
An additional challenge for the reproducibility of analysis workflows is the representation of statistical models across distinct implementations of analysis software. For example, GLM approaches to analyze fMRI time series are prevalent and supported by all of the major statistical packages (e.g., AFNI, SPM, FSL, Nilearn). However, specifying equivalent models across packages is non-trivial and requires time consuming package specific model specification [Pernet, 2014], which obfuscates details of the statistical model, exacerbates variability across pipelines, and makes it difficult to perform multiverse analyses (see Section 5.4). The BIDS Stats Model (BIDS-SM, see the resources table) specification has been proposed as a implementation-independent representation of fMRI GLM models; BIDS-SM describes the inputs, steps, and specification details of GLM-type analyses, and encodes them in a machine readable JSON format. The PyBIDS library provides tooling to facilitate reading BIDS-SM, and FitLins [Markiewicz et al., 2021] is a reference workflow that fits BIDS-SM using AFNI or Nilearn. The transformative potential of BIDS-SM is showcased by Neuroscout [de la Vega et al., 2022], a turnkey platform for fast and flexible neuroimaging analysis. Neuroscout provides a user-friendly web application for creating BIDS-SM on a curated set of public neuroimaging datasets, and leverages FitLins to fit statistical models in a fully reproducible and portable workflow. By standardizing the entire process of statistical modeling, users can formally specify a hypothesis and produce statistical results in a matter of minutes, while simultaneously ensuring a fully reproducible and transparent analysis that can be readily disseminated to the scientific community.
5.4 Multiverse analysis
The variety of data workflows reflects the enormous interest and the need for novel software instruments, but it also poses an important risk to reproducibility. The multitude of possible combinations of methods and parameters in each of the analysis steps creates an extremely large number of combinations to select from. This problem is often referred to as “researcher degrees of freedom” or “the garden of forking paths” [Gelman and Loken, 2013]. Importantly, analytical choices affect results. This has been shown for preprocessing of fMRI data already back in 2004 [Strother et al., 2004]. More work in this direction followed in 2012 [Churchill et al., 2012, Churchill et al., 2012]. While this work focused mainly on the aspect of tailoring preprocessing to e.g. maximize predictive models, recent efforts in fMRI (task fMRI: [Botvinik-Nezer et al., 2020, Carp, 2012]; preprocessing of resting-state fMRI: [Li et al., 2021]) and PET (specifically for preprocessing: [Nørgaard et al., 2020]) focused more on the variability of outcomes in general when analysis pipelines were varied. In addition, recent studies showed high variability in diffusion-based tractography dissection [Schilling et al., 2021] and event-related potentials in EEG preprocessing [Clayson et al., 2021, Šoškić et al., 2021]. Another large-scale attempt to estimate the analytical variability for EEG, EEGManyPipelines (see the resources table), is currently ongoing.
The converging findings of these studies across modalities suggest that it is important to test the robustness of reported results to specific analytical choices. One proposed solution to tackle the analytical variability, where many different analytical approaches are compared, is multiverse analysis [Hall et al., 2022]. There are two broad types of multiverse tools. In a “numerical instabilities” approach, different setups and numerical errors or uncertainties in computational tools are evaluated, analyses are rerun several times, and variability, robustness, and “mean answer” are estimated [Kiar et al., 2020]. One tool of this type that is being developed is “Fuzzy” [Kiar et al., 2021]. Alternatively, in a “classic multiverse analysis”, multiple pipelines are used with the same data and the results are compared across pipelines. Such an analysis could be conducted by a single or by multiple researchers [Aczel et al., 2021]. Although multiverse analysis was suggested before in other fields [Patel et al., 2015, Simonsohn et al., 2015, Steegen et al., 2016], there are not yet mature “classic multiverse analysis” tools for high-dimensional data like in neuroimaging. Explorable Multiverse Analyses is an R-tool that allows the readers to explore different statistical approaches in a paper [Dragicevic et al., 2019]. Other tools, such as the Python-based Boba [Liu et al., 2021], aim to facilitate multiverse analyses by allowing users to specify the shared and the varying parts of the code only once and by providing useful visualizations of the pipelines and results. However, these tools currently fit simpler analyses and datasets compared to the ones common in neuroimaging.
In neuroimaging, recent progress has been made in creating infrastructure for multiverse analysis in fMRI, based on the C-PAC tool (see Section 5.2; [Li et al., 2021]). Ongoing efforts to formalize machine-readable standards for statistical models (BIDS-SM) and pipelines to estimate them, and their integration with datasets using platforms such as brainlife.io [Avesani et al., 2019], could facilitate the development of multiverse tools. In order to make sense of a multiverse analysis, one needs methods to test for convergence across results of diverse analysis pipelines with the same data. Such a method for fMRI image-based meta-analysis was recently used in NARPS [Botvinik-Nezer et al., 2020] as well as in subsequent projects [Bowring et al., 2021]. Another simple statistical approach to a multiverse analysis was presented with PET data [Nørgaard et al., 2019], although it lacks statistical power, due to the use of a very conservative statistic. A different approach is to use active learning to approximate the whole multiverse space [Dafflon et al., 2020]. Moreover, Boos et al. {cite:t}{Boos2021-az} provided an online application to explore the effects of the choice of parameters on the results (data-driven auditory encoding, see the resources table). Progress is still needed until such tools are mature enough to allow scalable multiverse analysis in neuroimaging.
References on this page
- E1(1,2)
Alexandre Abraham, Fabian Pedregosa, Michael Eickenberg, Philippe Gervais, Andreas Mueller, Jean Kossaifi, Alexandre Gramfort, Bertrand Thirion, and Gaël Varoquaux. Machine learning for neuroimaging with scikit-learn. Frontiers in Neuroinformatics, 8:14, February 2014.
- E2
Balazs Aczel, Barnabas Szaszi, Gustav Nilsonne, Olmo R van den Akker, Casper J Albers, Marcel Alm van Assen, Jojanneke A Bastiaansen, Daniel Benjamin, Udo Boehm, Rotem Botvinik-Nezer, Laura F Bringmann, Niko A Busch, Emmanuel Caruyer, Andrea M Cataldo, Nelson Cowan, Andrew Delios, Noah Nn van Dongen, Chris Donkin, Johnny B van Doorn, Anna Dreber, Gilles Dutilh, Gary F Egan, Morton Ann Gernsbacher, Rink Hoekstra, Sabine Hoffmann, Felix Holzmeister, Juergen Huber, Magnus Johannesson, Kai J Jonas, Alexander T Kindel, Michael Kirchler, Yoram K Kunkels, D Stephen Lindsay, Jean-Francois Mangin, Dora Matzke, Marcus R Munafò, Ben R Newell, Brian A Nosek, Russell A Poldrack, Don van Ravenzwaaij, Jörg Rieskamp, Matthew J Salganik, Alexandra Sarafoglou, Tom Schonberg, Martin Schweinsberg, David Shanks, Raphael Silberzahn, Daniel J Simons, Barbara A Spellman, Samuel St-Jean, Jeffrey J Starns, Eric Luis Uhlmann, Jelte Wicherts, and Eric-Jan Wagenmakers. Consensus-based guidance for conducting and reporting multi-analyst studies. Elife, November 2021.
- E3
missing booktitle in Alfaro-Almagro2016-pj
- E4
Lau M Andersen. Group analysis in FieldTrip of Time-Frequency responses: a pipeline for reproducibility at every step of processing, going from individual sensor space representations to an Across-Group source space representation. Front. Neurosci., 12:261, May 2018.
- E5
Lau M Andersen. Group analysis in MNE-Python of evoked responses from a tactile stimulation paradigm: a pipeline for reproducibility at every step of processing, going from individual sensor space representations to an across-group source space representation. 2018.
- E6
Paolo Avesani, Brent McPherson, Soichi Hayashi, Cesar F Caiafa, Robert Henschel, Eleftherios Garyfallidis, Lindsey Kitchell, Daniel Bullock, Andrew Patterson, Emanuele Olivetti, Olaf Sporns, Andrew J Saykin, Lei Wang, Ivo Dinov, David Hancock, Bradley Caron, Yiming Qian, and Franco Pestilli. The open diffusion data derivatives, brain data upcycling via integrated publishing of derivatives and reproducible open cloud services. Scientific Data, 6(1):69, May 2019.
- E7(1,2)
Nick Barnes. Publish your computer code: it is good enough. Nature, 467(7317):753, October 2010.
- E8(1,2,3)
Rotem Botvinik-Nezer, Felix Holzmeister, Colin F Camerer, Anna Dreber, Juergen Huber, Magnus Johannesson, Michael Kirchler, Roni Iwanir, Jeanette A Mumford, R Alison Adcock, Paolo Avesani, Blazej M Baczkowski, Aahana Bajracharya, Leah Bakst, Sheryl Ball, Marco Barilari, Nadège Bault, Derek Beaton, Julia Beitner, Roland G Benoit, Ruud M W J Berkers, Jamil P Bhanji, Bharat B Biswal, Sebastian Bobadilla-Suarez, Tiago Bortolini, Katherine L Bottenhorn, Alexander Bowring, Senne Braem, Hayley R Brooks, Emily G Brudner, Cristian B Calderon, Julia A Camilleri, Jaime J Castrellon, Luca Cecchetti, Edna C Cieslik, Zachary J Cole, Olivier Collignon, Robert W Cox, William A Cunningham, Stefan Czoschke, Kamalaker Dadi, Charles P Davis, Alberto De Luca, Mauricio R Delgado, Lysia Demetriou, Jeffrey B Dennison, Xin Di, Erin W Dickie, Ekaterina Dobryakova, Claire L Donnat, Juergen Dukart, Niall W Duncan, Joke Durnez, Amr Eed, Simon B Eickhoff, Andrew Erhart, Laura Fontanesi, G Matthew Fricke, Shiguang Fu, Adriana Galván, Remi Gau, Sarah Genon, Tristan Glatard, Enrico Glerean, Jelle J Goeman, Sergej A E Golowin, Carlos González-García, Krzysztof J Gorgolewski, Cheryl L Grady, Mikella A Green, João F Guassi Moreira, Olivia Guest, Shabnam Hakimi, J Paul Hamilton, Roeland Hancock, Giacomo Handjaras, Bronson B Harry, Colin Hawco, Peer Herholz, Gabrielle Herman, Stephan Heunis, Felix Hoffstaedter, Jeremy Hogeveen, Susan Holmes, Chuan-Peng Hu, Scott A Huettel, Matthew E Hughes, Vittorio Iacovella, Alexandru D Iordan, Peder M Isager, Ayse I Isik, Andrew Jahn, Matthew R Johnson, Tom Johnstone, Michael J E Joseph, Anthony C Juliano, Joseph W Kable, Michalis Kassinopoulos, Cemal Koba, Xiang-Zhen Kong, Timothy R Koscik, Nuri Erkut Kucukboyaci, Brice A Kuhl, Sebastian Kupek, Angela R Laird, Claus Lamm, Robert Langner, Nina Lauharatanahirun, Hongmi Lee, Sangil Lee, Alexander Leemans, Andrea Leo, Elise Lesage, Flora Li, Monica Y C Li, Phui Cheng Lim, Evan N Lintz, Schuyler W Liphardt, Annabel B Losecaat Vermeer, Bradley C Love, Michael L Mack, Norberto Malpica, Theo Marins, Camille Maumet, Kelsey McDonald, Joseph T McGuire, Helena Melero, Adriana S Méndez Leal, Benjamin Meyer, Kristin N Meyer, Glad Mihai, Georgios D Mitsis, Jorge Moll, Dylan M Nielson, Gustav Nilsonne, Michael P Notter, Emanuele Olivetti, Adrian I Onicas, Paolo Papale, Kaustubh R Patil, Jonathan E Peelle, Alexandre Pérez, Doris Pischedda, Jean-Baptiste Poline, Yanina Prystauka, Shruti Ray, Patricia A Reuter-Lorenz, Richard C Reynolds, Emiliano Ricciardi, Jenny R Rieck, Anais M Rodriguez-Thompson, Anthony Romyn, Taylor Salo, Gregory R Samanez-Larkin, Emilio Sanz-Morales, Margaret L Schlichting, Douglas H Schultz, Qiang Shen, Margaret A Sheridan, Jennifer A Silvers, Kenny Skagerlund, Alec Smith, David V Smith, Peter Sokol-Hessner, Simon R Steinkamp, Sarah M Tashjian, Bertrand Thirion, John N Thorp, Gustav Tinghög, Loreen Tisdall, Steven H Tompson, Claudio Toro-Serey, Juan Jesus Torre Tresols, Leonardo Tozzi, Vuong Truong, Luca Turella, Anna E van 't Veer, Tom Verguts, Jean M Vettel, Sagana Vijayarajah, Khoi Vo, Matthew B Wall, Wouter D Weeda, Susanne Weis, David J White, David Wisniewski, Alba Xifra-Porxas, Emily A Yearling, Sangsuk Yoon, Rui Yuan, Kenneth S L Yuen, Lei Zhang, Xu Zhang, Joshua E Zosky, Thomas E Nichols, Russell A Poldrack, and Tom Schonberg. Variability in the analysis of a single neuroimaging dataset by many teams. Nature, 582(7810):84–88, June 2020.
- E9
missing note in Bowring2021-pw
- E10
Matthew Brett, Christopher J Markiewicz, Michael Hanke, Marc-Alexandre Côté, Ben Cipollini, Paul McCarthy, Dorota Jarecka, Christopher P Cheng, Yaroslav O Halchenko, Michiel Cottaar, Eric Larson, Satrajit Ghosh, Demian Wassermann, Stephan Gerhard, Gregory R Lee, Hao-Ting Wang, Erik Kastman, Jakub Kaczmarzyk, Roberto Guidotti, Or Duek, Jonathan Daniel, Ariel Rokem, Cindee Madison, Brendan Moloney, Félix C Morency, Mathias Goncalves, Ross Markello, Cameron Riddell, Christopher Burns, Jarrod Millman, Alexandre Gramfort, Jaakko Leppäkangas, Anibal Sólon, Jasper J F van den Bosch, Robert D Vincent, Henry Braun, Krish Subramaniam, Krzysztof J Gorgolewski, Pradeep Reddy Raamana, Julian Klug, B Nolan Nichols, Eric M Baker, Soichi Hayashi, Basile Pinsard, Christian Haselgrove, Mark Hymers, Oscar Esteban, Serge Koudoro, Fernando Pérez-García, Nikolaas N Oosterhof, Bago Amirbekian, Ian Nimmo-Smith, Ly Nguyen, Samir Reddigari, Samuel St-Jean, Egor Panfilov, Eleftherios Garyfallidis, Gael Varoquaux, Jon Haitz Legarreta, Kevin S Hahn, Oliver P Hinds, Bennet Fauber, Jean-Baptiste Poline, Jon Stutters, Kesshi Jordan, Matthew Cieslak, Miguel Estevan Moreno, Valentin Haenel, Yannick Schwartz, Zvi Baratz, Benjamin C Darwin, Bertrand Thirion, Carl Gauthier, Dimitri Papadopoulos Orfanos, Igor Solovey, Ivan Gonzalez, Jath Palasubramaniam, Justin Lecher, Katrin Leinweber, Konstantinos Raktivan, Markéta Calábková, Peter Fischer, Philippe Gervais, Syam Gadde, Thomas Ballinger, Thomas Roos, Venkateswara Reddy Reddam, and freec. Nipy/nibabel: 3.2.1. Zenodo, November 2020.
- E11(1,2,3)
Joshua Carp. On the plurality of (methodological) worlds: estimating the analytic flexibility of fmri experiments. Front. Neurosci., 6(OCT):1–13, 2012.
- E12(1,2)
Joshua Carp. The secret lives of experiments: methods reporting in the fMRI literature. Neuroimage, 63(1):289–300, October 2012.
- E13
Nathan W Churchill, Anita Oder, Hervé Abdi, Fred Tam, Wayne Lee, Christopher Thomas, Jon E Ween, Simon J Graham, and Stephen C Strother. Optimizing preprocessing and analysis pipelines for single-subject fMRI. i. standard temporal motion and physiological noise correction methods. Human Brain Mapping, 33(3):609–627, March 2012.
- E14
Nathan W Churchill, Grigori Yourganov, Anita Oder, Fred Tam, Simon J Graham, and Stephen C Strother. Optimizing preprocessing and analysis pipelines for single-subject fMRI: 2. interactions with ICA, PCA, task contrast and inter-subject heterogeneity. PLoS One, 7(2):e31147, February 2012.
- E15(1,2)
Peter E Clayson, Scott A Baldwin, Harold A Rocha, and Michael J Larson. The data-processing multiverse of event-related potentials (ERPs): a roadmap for the optimization and standardization of ERP processing and reduction pipelines. Neuroimage, pages 118712, November 2021.
- E16
R W Cox. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Computers and Biomedical Research, 29(29):162–173, 1996.
- E17
R W Cox and J S Hyde. Software tools for analysis and visualization of fMRI data. NMR in Biomedicine, 10(4-5):171–178, June 1997.
- E18
Cameron Craddock, Sharad Sikka, Brian Cheung, Ranjeet Khanuja, Satrajit S Ghosh, Chaogan Yan, Qingyang Li, Daniel Lurie, Joshua Vogelstein, Randal Burns, and Others. Towards automated analysis of connectomes: the configurable pipeline for the analysis of connectomes (c-pac). In Frontiers Neuroinformatics, volume 42, 10–3389. 2013.
- E19
missing note in Dafflon2020-cn
- E20
A M Dale, B Fischl, and M I Sereno. Cortical surface-based analysis. i. segmentation and surface reconstruction. Neuroimage, 9(2):179–194, February 1999.
- E21
A M Dale and Martin I Sereno. Improved localizadon of cortical activity by combining EEG and MEG with MRI cortical surface reconstruction: a linear approach. Journal of Cognitive Neuroscience, 5(2):162–176, 1993.
- E22
missing note in De_la_Vega2022-xo
- E23
Arnaud Delorme and Scott Makeig. EEGLAB: an open source toolbox for analysis of single-trial EEG dynamics including independent component analysis. Journal of Neuroscience Methods, 134(1):9–21, March 2004.
- E24
missing booktitle in Delorme2021-ue
- E25
J A Desjardins, S van Noordt, S Huberty, S J Segalowitz, and M Elsabbagh. EEG integrated platform lossless (EEG-IP-L) pre-processing pipeline for objective signal quality assessment incorporating data annotation and blind source separation. Journal of Neuroscience Methods, 347:108961, January 2021.
- E26
missing booktitle in Dragicevic2019-mj
- E27
Oscar Esteban, Rastko Ciric, Karolina Finc, Ross W Blair, Christopher J Markiewicz, Craig A Moodie, James D Kent, Mathias Goncalves, Elizabeth DuPre, Daniel E P Gomez, Zhifang Ye, Taylor Salo, Romain Valabregue, Inge K Amlien, Franziskus Liem, Nir Jacoby, Hrvoje Stojić, Matthew Cieslak, Sebastian Urchs, Yaroslav O Halchenko, Satrajit S Ghosh, Alejandro De La Vega, Tal Yarkoni, Jessey Wright, William H Thompson, Russell A Poldrack, and Krzysztof J Gorgolewski. Analysis of task-based functional MRI data preprocessed with fMRIPrep. Nature Protocols, 15(7):2186–2202, July 2020.
- E28
Oscar Esteban, Christopher J Markiewicz, Ross W Blair, Craig A Moodie, A Ilkay Isik, Asier Erramuzpe, James D Kent, Mathias Goncalves, Elizabeth DuPre, Madeleine Snyder, Hiroyuki Oya, Satrajit S Ghosh, Jessey Wright, Joke Durnez, Russell A Poldrack, and Krzysztof J Gorgolewski. fMRIPrep: a robust preprocessing pipeline for functional MRI. Nature Methods, 16(1):111–116, January 2019.
- E29(1,2)
Guillaume Flandin and Karl Friston. Statistical parametric mapping (SPM). Scholarpedia J., 3(4):6232, 2008.
- E30
A Gelman and E Loken. The garden of forking paths: why multiple comparisons can be a problem, even when there is no `fishing expedition' or `p-hacking' and the research hypothesis was posited ahead of time. http://www.stat.columbia.edu/ gelman/research/unpublished/p_hacking.pdf, 2013. Accessed: NA-NA-NA.
- E31
Matthew F Glasser, Stamatios N Sotiropoulos, J Anthony Wilson, Timothy S Coalson, Bruce Fischl, Jesper L Andersson, Junqian Xu, Saad Jbabdi, Matthew Webster, Jonathan R Polimeni, David C Van Essen, Mark Jenkinson, and WU-Minn HCP Consortium. The minimal preprocessing pipelines for the human connectome project. Neuroimage, 80:105–124, October 2013.
- E32
Krzysztof J Gorgolewski, Christopher D Burns, Cindee Madison, Dav Clark, Yaroslav O Halchenko, Michael L Waskom, and Satrajit S Ghosh. Nipype: a flexible, lightweight and extensible neuroimaging data processing framework in python. Frontiers in Neuroinformatics, 2011.
- E33(1,2)
Krzysztof J Gorgolewski and Russell A Poldrack. A practical guide for improving transparency and reproducibility in neuroimaging research. PLoS Biology, 14(7):e1002506, July 2016.
- E34
Alexandre Gramfort, Martin Luessi, Eric Larson, Denis A Engemann, Daniel Strohmeier, Christian Brodbeck, Roman Goj, Mainak Jas, Teon Brooks, Lauri Parkkonen, and Matti Hämäläinen. MEG and EEG data analysis with MNE-Python. Frontiers in Neuroscience, 7:267, December 2013.
- E35
Alexandre Gramfort, Martin Luessi, Eric Larson, Denis A Engemann, Daniel Strohmeier, Christian Brodbeck, Lauri Parkkonen, and Matti S Hämäläinen. MNE software for processing MEG and EEG data. Neuroimage, 86:446–460, February 2014.
- E36
Brian D Hall, Yang Liu, Yvonne Jansen, Pierre Dragicevic, Fanny Chevalier, and Matthew Kay. A survey of tasks and visualizations in multiverse analysis reports. Computer Graphics Forum, February 2022.
- E37
Richard N Henson, Hunar Abdulrahman, Guillaume Flandin, and Vladimir Litvak. Multimodal integration of M/EEG and f/MRI data in SPM12. Front. Neurosci., 13:300, April 2019.
- E38(1,2)
Mainak Jas, Eric Larson, Denis A Engemann, Jaakko Leppäkangas, Samu Taulu, Matti Hämäläinen, and Alexandre Gramfort. A reproducible MEG/EEG group study with the MNE software: recommendations, quality assessments, and good practices. Frontiers in Neuroscience, 12:530, August 2018.
- E39
Mark Jenkinson, Christian F Beckmann, Timothy E J Behrens, Mark W Woolrich, and Stephen M Smith. FSL. Neuroimage, 62(2):782–790, August 2012.
- E40
Greg Kiar, Yohan Chatelain, Tristan Glatard, Ali Salari, Pablo de Oliveira Castro, michaelnicht, Antoine Hébert, and Mayank Vadariya. Verificarlo/fuzzy: fuzzy v0.5.0. June 2021.
- E41
Gregory Kiar, Pablo de Oliveira Castro, Pierre Rioux, Eric Petit, Shawn T Brown, Alan C Evans, and Tristan Glatard. Comparing perturbation models for evaluating stability of neuroimaging pipelines. The International Journal of High Performance Computing Applications, 34(5):491–501, September 2020.
- E42(1,2)
X Li, L Ai, S Giavasis, H Jin, E Feczko, T Xu, J Clucas, A Franco, A S Heinsfeld, A Adebimpe, and Others. Moving beyond processing and analysis-related variation in neuroscience. bioRxiv, 2021.
- E43
Vladimir Litvak, Jérémie Mattout, Stefan Kiebel, Christophe Phillips, Richard Henson, James Kilner, Gareth Barnes, Robert Oostenveld, Jean Daunizeau, Guillaume Flandin, Will Penny, and Karl Friston. EEG and MEG data analysis in SPM8. Computational Intelligence and Neuroscience, 2011:852961, March 2011.
- E44
Yang Liu, Alex Kale, Tim Althoff, and Jeffrey Heer. Boba: authoring and visualizing multiverse analyses. IEEE Transactions on Visualization and Computer Graphics, 27(2):1753–1763, February 2021.
- E45
Christopher J Markiewicz, Alejandro De La Vega, Adina Wagner, Yaroslav O Halchenko, Karolina Finc, Rastko Ciric, Mathias Goncalves, Dylan M Nielson, James D Kent, John A Lee, Russell A Poldrack, and Krzysztof J Gorgolewski. Poldracklab/fitlins: v0.9.2. July 2021.
- E46
Marlene Meyer, Didi Lamers, Ezgi Kayhan, Sabine Hunnius, and Robert Oostenveld. Enhancing reproducibility in developmental EEG research: BIDS, cluster-based permutation tests, and effect sizes. Dev. Cogn. Neurosci., 52:101036, December 2021.
- E47
Guiomar Niso, Laurens R Krol, Etienne Combrisson, A-Sophie Dubarry, Madison A Elliott, Clément François, Yseult Héjja-Brichard, Sophie K Herbst, Karim Jerbi, Vanja Kovic, Katia Lehongre, Steven J Luck, Manuel Mercier, John C Mosher, Yuri G Pavlov, Aina Puce, Antonio Schettino, Daniele Schön, Walter Sinnott-Armstrong, Bertille Somon, Anđela Šoškić, Suzy J Styles, Roni Tibon, Martina G Vilas, Marijn van Vliet, and Maximilien Chaumon. Good scientific practice in MEEG research: progress and perspectives. Neuroimage, pages 119056, March 2022.
- E48
Guiomar Niso, Francois Tadel, Elizabeth Bock, Martin Cousineau, Andrés Santos, and Sylvain Baillet. Brainstorm pipeline analysis of Resting-State data from the open MEG archive. Front. Neurosci., 13:284, April 2019.
- E49(1,2)
Martin Nørgaard, Melanie Ganz, Claus Svarer, Vibe G Frokjaer, Douglas N Greve, Stephen C Strother, and Gitte M Knudsen. Different preprocessing strategies lead to different conclusions: a [11C]DASB-PET reproducibility study. Journal of Cerebral Blood Flow and Metabolism, 40(9):1902–1911, September 2020.
- E50
missing booktitle in Norgaard2019-su
- E51
Robert Oostenveld, Pascal Fries, Eric Maris, and Jan-Mathijs Schoffelen. FieldTrip: open source software for advanced analysis of MEG, EEG, and invasive electrophysiological data. Computational Intelligence and Neuroscience, 2011:156869, 2011.
- E52
Chirag J Patel, Belinda Burford, and John P A Ioannidis. Assessment of vibration of effects due to model specification can demonstrate the instability of observational associations. Journal of Clinical Epidemiology, 68(9):1046–1058, September 2015.
- E53
William D Penny, Karl J Friston, John T Ashburner, Stefan J Kiebel, and Thomas E Nichols. Statistical Parametric Mapping: The analysis of functional brain images. Elsevier, April 2011.
- E54
Cyril R Pernet. Misconceptions in the use of the general linear model applied to functional MRI: a tutorial for junior neuro-imagers. Front. Neurosci., 8:1, January 2014.
- E55
Cyril R Pernet, Ramon Martinez-Cancino, Dung Truong, Scott Makeig, and Arnaud Delorme. From BIDS-Formatted EEG data to Sensor-Space group results: a fully reproducible workflow with EEGLAB and LIMO EEG. Front. Neurosci., 14:610388, 2020.
- E56
Tzvetan Popov, Robert Oostenveld, and Jan M Schoffelen. FieldTrip made easy: an analysis protocol for group analysis of the auditory steady state brain response in time, frequency, and space. Front. Neurosci., 12:711, October 2018.
- E57(1,2)
Kurt G Schilling, François Rheault, Laurent Petit, Colin B Hansen, Vishwesh Nath, Fang-Cheng Yeh, Gabriel Girard, Muhamed Barakovic, Jonathan Rafael-Patino, Thomas Yu, Elda Fischi-Gomez, Marco Pizzolato, Mario Ocampo-Pineda, Simona Schiavi, Erick J Canales-Rodríguez, Alessandro Daducci, Cristina Granziera, Giorgio Innocenti, Jean-Philippe Thiran, Laura Mancini, Stephen Wastling, Sirio Cocozza, Maria Petracca, Giuseppe Pontillo, Matteo Mancini, Sjoerd B Vos, Vejay N Vakharia, John S Duncan, Helena Melero, Lidia Manzanedo, Emilio Sanz-Morales, Ángel Peña-Melián, Fernando Calamante, Arnaud Attyé, Ryan P Cabeen, Laura Korobova, Arthur W Toga, Anupa Ambili Vijayakumari, Drew Parker, Ragini Verma, Ahmed Radwan, Stefan Sunaert, Louise Emsell, Alberto De Luca, Alexander Leemans, Claude J Bajada, Hamied Haroon, Hojjatollah Azadbakht, Maxime Chamberland, Sila Genc, Chantal M W Tax, Ping-Hong Yeh, Rujirutana Srikanchana, Colin D Mcknight, Joseph Yuan-Mou Yang, Jian Chen, Claire E Kelly, Chun-Hung Yeh, Jerome Cochereau, Jerome J Maller, Thomas Welton, Fabien Almairac, Kiran K Seunarine, Chris A Clark, Fan Zhang, Nikos Makris, Alexandra Golby, Yogesh Rathi, Lauren J O'Donnell, Yihao Xia, Dogu Baran Aydogan, Yonggang Shi, Francisco Guerreiro Fernandes, Mathijs Raemaekers, Shaun Warrington, Stijn Michielse, Alonso Ramírez-Manzanares, Luis Concha, Ramón Aranda, Mariano Rivera Meraz, Garikoitz Lerma-Usabiaga, Lucas Roitman, Lucius S Fekonja, Navona Calarco, Michael Joseph, Hajer Nakua, Aristotle N Voineskos, Philippe Karan, Gabrielle Grenier, Jon Haitz Legarreta, Nagesh Adluru, Veena A Nair, Vivek Prabhakaran, Andrew L Alexander, Koji Kamagata, Yuya Saito, Wataru Uchida, Christina Andica, Masahiro Abe, Roza G Bayrak, Claudia A M Gandini Wheeler-Kingshott, Egidio D'Angelo, Fulvia Palesi, Giovanni Savini, Nicolò Rolandi, Pamela Guevara, Josselin Houenou, Narciso López-López, Jean-François Mangin, Cyril Poupon, Claudio Román, Andrea Vázquez, Chiara Maffei, Mavilde Arantes, José Paulo Andrade, Susana Maria Silva, Vince D Calhoun, Eduardo Caverzasi, Simone Sacco, Michael Lauricella, Franco Pestilli, Daniel Bullock, Yang Zhan, Edith Brignoni-Perez, Catherine Lebel, Jess E Reynolds, Igor Nestrasil, René Labounek, Christophe Lenglet, Amy Paulson, Stefania Aulicka, Sarah R Heilbronner, Katja Heuer, Bramsh Qamar Chandio, Javier Guaje, Wei Tang, Eleftherios Garyfallidis, Rajikha Raja, Adam W Anderson, Bennett A Landman, and Maxime Descoteaux. Tractography dissection variability: what happens when 42 groups dissect 14 white matter bundles on the same dataset? Neuroimage, 243:118502, November 2021.
- E58
Uri Simonsohn, Joseph P Simmons, and Leif D Nelson. Specification curve: descriptive and inferential statistics on all reasonable specifications. SSRN Electronic Journal, 2015.
- E59
Sara Steegen, Francis Tuerlinckx, Andrew Gelman, and Wolf Vanpaemel. Increasing transparency through a multiverse analysis. Perspectives on Psychological Science, 11(5):702–712, 2016.
- E60
Stephen C Strother, Stephen La Conte, Lars Kai Hansen, Jon Anderson, Jin Zhang, Sujit Pulapura, and David Rottenberg. Optimizing the fMRI data-processing pipeline using prediction and reproducibility performance metrics: i. a preliminary group analysis. Neuroimage, 23 Suppl 1:S196–207, 2004.
- E61
François Tadel, Sylvain Baillet, John C Mosher, Dimitrios Pantazis, and Richard M Leahy. Brainstorm: a user-friendly application for MEG/EEG analysis. Computational Intelligence and Neuroscience, 2011:879716, April 2011.
- E62(1,2)
François Tadel, Elizabeth Bock, Guiomar Niso, John C Mosher, Martin Cousineau, Dimitrios Pantazis, Richard M Leahy, and Sylvain Baillet. MEG/EEG group analysis with brainstorm. Frontiers in Neuroscience, 13:76, February 2019.
- E63
Nicholas J Tustison, Hans J Johnson, Torsten Rohlfing, Arno Klein, Satrajit S Ghosh, Luis Ibanez, and Brian B Avants. Instrumentation bias in the use and evaluation of scientific software: recommendations for reproducible practices in the computational sciences. Frontiers in Neuroscience, 7:162, September 2013.
- E64
Marijn van Vliet, Mia Liljeström, Susanna Aro, Riitta Salmelin, and Jan Kujala. Analysis of functional connectivity and oscillatory power using DICS: from raw MEG data to Group-Level statistics in python. Front. Neurosci., 12:586, September 2018.
- E65(1,2)
Anđela Šoškić, Vojislav Jovanović, Suzy J Styles, Emily S Kappenman, and Vanja Ković. How to do better N400 studies: reproducibility, consistency and adherence to research standards in the existing literature. Neuropsychology Review, 2021.
- 1
Sources: Icons from the Noun Project: Software by Adrien Coquet; Workflow by D. Sahua; Statistics by Creative Stall; Chaos Sigil by Avana Vana; Logos: used with permission by the copyright holders.