Research data management
Research data management#
Issue
Good research data management (RDM), i.e. how data are organized, maintained, annotated, tracked, stored, and accessed throughout a research project, forms the basic foundations of result reproducibility, data reusability, and research efficiency [Gorgolewski and Poldrack, 2016, Nosek et al., 2012, Poldrack et al., 2017, Poldrack et al., 2019, Poldrack et al., 2020, Wilkinson et al., 2016, Nosek et al., 2018, Borghi and Van Gulick, 2021, Nosek and Lakens, 2014, Poline et al., 2022]. Consequently, Data Management Plans (DMPs) are widely required by funders even at the application phase (e.g., NIH and NSF in the U.S., ERC in Europe), increasingly expected by scientific peers, and holds considerable benefits for individual researchers.
What do we provide
It is good practice to develop, review and execute DMPs for every experiment, whether or not it is required by the funding agency. While specific RDM requirements vary across subdisciplines, this section highlights RDM standards and tools applicable across neuroimaging, ranging from data organization to annotation and publication.
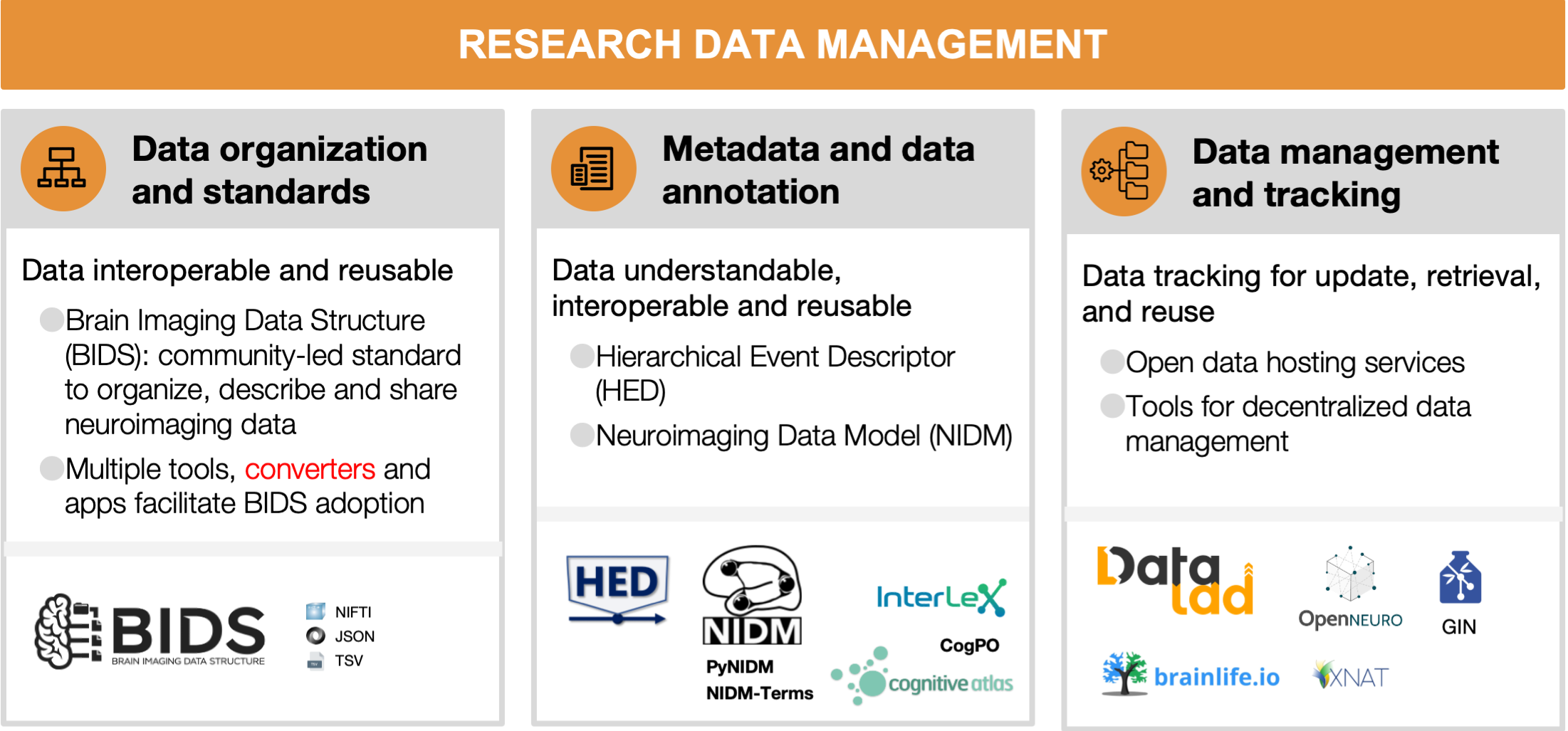
4.1 Data organization and standards
Neuroimaging experiments result in complicated data that can be arranged in many different ways. Historically, data were organized differently between institutions and within labs. This lack of consensus (or a standard) could lead to misunderstandings and suboptimal usage of various resources: human (e.g., time wasted on rearranging data or rewriting scripts expecting certain structure), infrastructure (e.g., data storage space, duplicates), and financial (e.g., disorganized data has limited longevity and value after first publication, because it is hard or even impossible for other researchers to understand and use it). Finally, and most importantly, it produces poor reproducibility of results, even within the lab where data were collected, because it is more likely to include errors and less likely to be accessible to future lab members (or even to the original researcher who obtained the data, months or years after they worked on it). Therefore, the need for a data standard in the neuroimaging community became essential.
The Brain Imaging Data Structure (BIDS) is a community-led standard for organizing, describing, and sharing neuroimaging data [RRID:SCR_016124]. BIDS is an evolving standard, which supports multiple neuroimaging modalities including MRI [Gorgolewski et al., 2016], MEG [Niso et al., 2018], EEG [Pernet et al., 2019], intracranial EEG [Holdgraf et al., 2019], PET [Nørgaard et al., 2019], Microscopy [Bourget et al., 2022], and imaging genetics [Moreau et al., 2020]. Many more extensions are under active development, for example, fNIRS, motion capture, and animal neurophysiology. The BIDS specification documents how to organize the data, generally based on simple file formats (such as NIfTI for tomographic data [Cox et al., 2004], and JSON for metadata) and folder structures. This specification can be extended through community-driven processes to incorporate new neuroimaging modalities or sets of data types.
Multiple applications and tools have been released to make it easy for researchers to incorporate BIDS into their current workflows, maximizing reproducibility, enabling effective data sharing, and supporting good data management practices. For example, BIDS converters make it easier to convert data into BIDS format (e.g., MNE-BIDS [Appelhoff et al., 2019] for MEG and EEG, dcm2bids, ReproNim’s HeuDiConv [Halchenko et al., 2021] and ReproIn [Visconti di Oleggio Castello et al., 2020] for MRI and PET2BIDS for PET; see many more on the resources table). The BIDS validator can help researchers make sure their dataset is BIDS-valid following conversion.
Once data are in BIDS, tools are available to ease interaction with the data. Two commonly used software packages are PyBIDS {cite:p}{Yarkoni2019-zm}, and BIDS-Matlab [Gau et al., 2022]. These tools facilitate useful dataset queries—such as how many participants are part of a dataset or what tasks were performed— as well as programmatically retrieving specific files—such as all functional runs for a specific subject. Finally, BIDS apps are containerized analysis pipelines that use full BIDS datasets as their input and produce derivative data [Gorgolewski et al., 2017]. Examples of BIDS apps include MRIQC [Esteban et al., 2017], for MRI quality control, fMRIPrep [Esteban et al., 2019] for fMRI preprocessing, and PyMVPA [Hanke et al., 2009] for statistical learning analyses of large datasets (see more at the resources table).
BIDS is a community-led standard and strives to be open and inclusive. The BIDS specification is the result of the ongoing collaboration, shared knowledge, discussion, and consensus through the email discussion list, shared Google docs, and GitHub. Questions are also answered on the Neurostars forum and the Brainhack Mattermost channel. BIDS has a well-specified governance structure where everybody is welcome to participate (Code of Conduct), and the BIDS Starter Kit is a growing resource intended to simplify the learning process for newcomers.
4.2 Metadata and data annotation
Metadata and data annotation induce consistency and facilitate data replication and reuse. It improves the clarity of the dataset, the ability for collaborators to understand the conditions in which the data were collected, and the ability to effectively share and reuse them. Commonly, metadata files are data dictionaries that map key terms from an agreed-upon vocabulary to data values that contain detailed and standardized information about the key terms. For example, a key called “SampleFrequency” might map to a numerical value, or a key “TaskDescription” might map to a free-form text that describes the task used in a specific experiment. The BIDS standard has proposed a consistent metadata structure in its specification along with a set of specification terms and tags.
Data annotation is also crucial for most data analyses in neuroimaging. For example, when analyzing task-based data, an experiment’s reproducibility is largely determined by the extent to which events are clearly documented. Beyond reproducing previous findings, exhaustively annotated events can allow researchers to re-use the data for means that were originally not thought of at data collection time [Bigdely-Shamlo et al., 2020]. However, even if each study is fully annotated, without a standard to consistently describe facets of events, all annotations will remain cumbersome and error-prone to work with, and achieving a state of machine readability will require effortful labor.
To address this problem, the Hierarchical Event Descriptor (HED) standard has been continuously developed over the past years [Robbins et al., 2021, Robbins et al., 2021]. Drawing on a set of hierarchical vocabulary structures (the HED base schema) and application rules, the HED standard allows for both human- and machine readability, validation, and search of annotations across studies. HED is furthermore fully integrated with the BIDS standard (see Section 4.1), and can be extended by researcher supplied schemas.
Additionally, the Neuroimaging Data Model [Keator et al., 2013, Maumet et al., 2016] effort aims to build a core structure for neuroscience datasets to improve searching across publicly-available datasets. The initiative also provides tools to create and use NIDM documents from BIDS datasets [Appelhoff et al., 2019]. To effectively describe neuroscience data, well-developed community-driven vocabularies are needed. NIDM is built using semantic web techniques and builds off the PROV (provenance) vocabulary [Moreau et al., 2015]. Moreover, the NIDM-Terms effort has begun to collect and extend sets of community-developed controlled vocabularies and techniques for associating concepts with selected study variables of publicly-available neuroimaging datasets (i.e., OpenNeuro, ABIDE, ADHD200, and CoRR). This keeps a registry of the domain-relevant vocabularies and concepts used to annotate datasets, further facilitating concept reuse, and improved inter-dataset search. The NIDM team has developed a JavaScript web application, as well as Python-based command line annotation tools, that allow researchers to annotate their BIDS structured datasets and single tabular files (e.g., csv and tsv spreadsheets), and export BIDS JSON-formatted data dictionaries, NIDM JSON-LD data dictionaries, and NIDM semantic web documents, into sidecar files that accompany the data files. Currently, the NIDM-Terms annotation tools allow researchers to associate their study variables with concepts available in the Cognitive Atlas [Poldrack et al., 2011], the InterLex information resource, and those in the canonical NIDM terminology/ontology as well as encourage them to add descriptive information to improve the clarity of their variables. Such an effort harmonizes and improves the consistency of neuroimaging data and thus makes querying across neuroimaging datasets more efficient.
4.3 Data management and tracking
Raw data and derivatives (outputs from processed data) form the basis for scientific analyses and insights. Being able to efficiently store, retrieve, and update data, derivatives, and metadata across a variety of available storage options is crucial to enable further research [Borghi and Van Gulick, 2021]. As files change and evolve over the course of a project, there is a need to identify which data have been used in the generation of a result, and, in case the data were subject to change or updates, which exact version of the data has been used. The ability to manage data and metadata and track the data-analysis process provides a basis for rigor and reproducibility.
DataLad [Halchenko et al., 2021] is an open-source, community-developed, general purpose tool for managing and version controlling digital files in a decentralized manner. It tracks data of any type or size in a scalable, Git-repository-based overlay structure, called the dataset (practically, a structure of folders and files). DataLad allows tracking data and metadata files stored on local devices as well as remote or cloud infrastructure. DataLad can retrieve public data from major providers such as OpenNeuro, the Canadian Open Neuroscience Platform, the International Neuroimaging Data-sharing Initiative, the Healthy Brain Network Serial Scanning Initiative, Data sharing for Collaborative Research in Computational Neuroscience, the Human Connectome Project’s open access dataset [Van Essen et al., 2013], and many more. Beyond public data, with appropriate permissions or authentication, it can retrieve data from web-based storage providers including major cloud storage services, and local and remote paths [Halchenko et al., 2021, Hanke et al., 2021]. DataLad implements this decentralized data management functionality in order to ensure streamlined access to tracked data regardless of hosting service, and to expose datasets for easy access on repository hosting structure. It separates management of file content from lean metadata management by tracking pointers to the services that host managed files (i.e., local infrastructure, remote hosting services, or multiple storage solutions at once). Using these pointers, it enables streamlined on-demand file retrieval in uniquely identified versions from the registered source. Importantly, data retrieval works via streamlined commands regardless of where the data are hosted. Information about DataLad can be found in the DataLad Handbook ([Wagner et al., 2021], see the resources table). Entire computing environments could be efficiently managed in DataLad using datalad-container extension [Meyer et al., 2021] developed in collaboration between DataLad and ReproNim projects.
Brainlife.io is another open science project that allows data management. Brainlife.io is a free and open community-oriented, non-commercial cloud platform that provides web services to support reproducible data management and analysis. Brainlife.io tracks data provenance automatically for the users. As data are analyzed using the Graphical User Interfaces (GUI) and the platform’s data processing applications, provenance metadata information is automatically generated and stored associated with the data derivatives. The users do not have to manually save data versions, the platform does that automatically and it allows visualizing data provenance graphs.
DataLad and brainlife.io are synergistic but not overlapping projects that address different user bases and needs. Indeed, DataLad and Brainlife.io interact nicely with one another and all published datasets retrieved by DataLad are readily accessible at brainlife.io datasets.
References on this page
- D1
Stefan Appelhoff, Julianna F Bates, Satrajit Ghosh, David B Keator, David N Kennedy, Russell Poldrack, Jean-Baptiste Poline, Jason Steffener, B Nolan Nichols, Franklin Feingold, Cyril Pernet, Gustav Nilsonne, Camille Maumet, Guillaume Flandin, Rémi Gau, Robert Oostenveld, Elizabeth Dupré, Arnaud Delorme, Christopher J Markiewicz, Natacha Perez, Karl G Helmer, Dorota Jarecka, Jeffrey S Grethe, Dianne Patterson, Tibor Auer, Hauke Bartsch, Thomas E Nichols, Vince Calhoun, Melanie Ganz, Robert E Smith, and Tal Yarkoni. BIDS and the NeuroImaging data model (NIDM). F1000Research, 8(1924):1924, 2019.
- D2
Stefan Appelhoff, Matthew Sanderson, Teon Brooks, Marijn van Vliet, Romain Quentin, Chris Holdgraf, Maximilien Chaumon, Ezequiel Mikulan, Kambiz Tavabi, Richard Höchenberger, Dominik Welke, Clemens Brunner, Alexander Rockhill, Eric Larson, Alexandre Gramfort, and Mainak Jas. MNE-BIDS: organizing electrophysiological data into the BIDS format and facilitating their analysis. Journal of Open Source Software, 4(44):1896, 2019.
- D3
Nima Bigdely-Shamlo, Jonathan Touryan, Alejandro Ojeda, Christian Kothe, Tim Mullen, and Kay Robbins. Automated EEG mega-analysis i: spectral and amplitude characteristics across studies. Neuroimage, 207:116361, February 2020.
- D4
missing note in Borghi2021-go
- D5
John A Borghi and Ana E Van Gulick. Data management and sharing: practices and perceptions of psychology researchers. PLoS One, 16(5):e0252047, May 2021.
- D6
Marie-Hélène Bourget, Lee Kamentsky, Satrajit S Ghosh, Giacomo Mazzamuto, Alberto Lazari, Christopher J Markiewicz, Robert Oostenveld, Guiomar Niso, Yaroslav O Halchenko, and Ilona Lipp. Microscopy-bids: an extension to the brain imaging data structure for microscopy data. Frontiers in Neuroscience, 2022.
- D7
missing booktitle in Cox2004-rn
- D8
Oscar Esteban, Daniel Birman, Marie Schaer, Oluwasanmi O Koyejo, Russell A Poldrack, and Krzysztof J Gorgolewski. MRIQC: advancing the automatic prediction of image quality in MRI from unseen sites. PLoS One, 12(9):e0184661, September 2017.
- D9
Oscar Esteban, Christopher J Markiewicz, Ross W Blair, Craig A Moodie, A Ilkay Isik, Asier Erramuzpe, James D Kent, Mathias Goncalves, Elizabeth DuPre, Madeleine Snyder, Hiroyuki Oya, Satrajit S Ghosh, Jessey Wright, Joke Durnez, Russell A Poldrack, and Krzysztof J Gorgolewski. fMRIPrep: a robust preprocessing pipeline for functional MRI. Nature Methods, 16(1):111–116, January 2019.
- D10
Rémi Gau, Guillaume Flandin, Andrew Janke, tanguyduval, Robert Oostenveld, Christopher Madan, Guiomar Niso Galán, Michał Szczepanik, Henk Mutsaerts, Nikita Beliy, Martin Norgaard, Cyril Pernet, and Phillips Chrisophe. Bids-matlab. January 2022.
- D11
Krzysztof J Gorgolewski, Fidel Alfaro-Almagro, Tibor Auer, Pierre Bellec, Mihai Capotă, M Mallar Chakravarty, Nathan W Churchill, Alexander Li Cohen, R Cameron Craddock, Gabriel A Devenyi, Anders Eklund, Oscar Esteban, Guillaume Flandin, Satrajit S Ghosh, J Swaroop Guntupalli, Mark Jenkinson, Anisha Keshavan, Gregory Kiar, Franziskus Liem, Pradeep Reddy Raamana, David Raffelt, Christopher J Steele, Pierre-Olivier Quirion, Robert E Smith, Stephen C Strother, Gaël Varoquaux, Yida Wang, Tal Yarkoni, and Russell A Poldrack. BIDS apps: improving ease of use, accessibility, and reproducibility of neuroimaging data analysis methods. PLoS Computational Biology, 13(3):e1005209, March 2017.
- D12
Krzysztof J Gorgolewski, Tibor Auer, Vince D Calhoun, R Cameron Craddock, Samir Das, Eugene P Duff, Guillaume Flandin, Satrajit S Ghosh, Tristan Glatard, Yaroslav O Halchenko, Daniel A Handwerker, Michael Hanke, David Keator, Xiangrui Li, Zachary Michael, Camille Maumet, B Nolan Nichols, Thomas E Nichols, John Pellman, Jean Baptiste Poline, Ariel Rokem, Gunnar Schaefer, Vanessa Sochat, William Triplett, Jessica A Turner, Gaël Varoquaux, and Russell A Poldrack. The brain imaging data structure: a format for organizing and describing outputs of neuroimaging experiments. Scientific Data, 3:1–9, 2016.
- D13
Krzysztof J Gorgolewski and Russell A Poldrack. A practical guide for improving transparency and reproducibility in neuroimaging research. PLoS Biology, 14(7):e1002506, July 2016.
- D14
Yaroslav Halchenko, Mathias Goncalves, Matteo Visconti di Oleggio Castello, Satrajit Ghosh, Taylor Salo, Michael Hanke, Pablo Velasco, Dae, James Kent, Matthew Brett, Inge Amlien, Chris Gorgolewski, Darren Christopher Lukas, Chris Markiewicz, Steven Tilley, Jakub Kaczmarzyk, Joerg Stadler, Sin Kim, Ari Kahn, Benjamin Poldrack, Bruno Melo, Henry Braun, John Pellman, Daniel Lurie, John Lee, Adina Wagner, Franklin Feingold, Johan Carlin, Kalle Samuels, and Kyle Meyer. Nipy/heudiconv:. October 2021.
- D15(1,2)
Yaroslav Halchenko, Kyle Meyer, Benjamin Poldrack, Debanjum Solanky, Adina Wagner, Jason Gors, Dave MacFarlane, Dorian Pustina, Vanessa Sochat, Satrajit Ghosh, Christian Mönch, Christopher Markiewicz, Laura Waite, Ilya Shlyakhter, Alejandro de la Vega, Soichi Hayashi, Christian Häusler, Jean-Baptiste Poline, Tobias Kadelka, Kusti Skytén, Dorota Jarecka, David Kennedy, Ted Strauss, Matt Cieslak, Peter Vavra, Horea-Ioan Ioanas, Robin Schneider, Mika Pflüger, James Haxby, Simon Eickhoff, and Michael Hanke. DataLad: distributed system for joint management of code, data, and their relationship. Journal of Open Source Software, 6(63):3262, 2021.
- D16
Michael Hanke, Yaroslav O Halchenko, Per B Sederberg, Stephen José Hanson, James V Haxby, and Stefan Pollmann. PyMVPA: a python toolbox for multivariate pattern analysis of fMRI data. Neuroinformatics, 7(1):37–53, January 2009.
- D17
Michael Hanke, Franco Pestilli, Adina S Wagner, Christopher J Markiewicz, Jean-Baptiste Poline, and Yaroslav O Halchenko. In defense of decentralized research data management. Neuroforum, 27(1):17–25, February 2021.
- D18
Christopher Holdgraf, Stefan Appelhoff, Stephan Bickel, Kristofer Bouchard, Sasha D'Ambrosio, Olivier David, Orrin Devinsky, Benjamin Dichter, Adeen Flinker, Brett L Foster, Krzysztof J Gorgolewski, Iris Groen, David Groppe, Aysegul Gunduz, Liberty Hamilton, Christopher J Honey, Mainak Jas, Robert Knight, Jean-Philippe Lachaux, Jonathan C Lau, Christopher Lee-Messer, Brian N Lundstrom, Kai J Miller, Jeffrey G Ojemann, Robert Oostenveld, Natalia Petridou, Gio Piantoni, Andrea Pigorini, Nader Pouratian, Nick F Ramsey, Arjen Stolk, Nicole C Swann, François Tadel, Bradley Voytek, Brian A Wandell, Jonathan Winawer, Kirstie Whitaker, Lyuba Zehl, and Dora Hermes. iEEG-BIDS: extending the brain imaging data structure specification to human intracranial electrophysiology. Scientific Data, 6(1):102, June 2019.
- D19
D B Keator, K Helmer, J Steffener, J A Turner, T G M Van Erp, S Gadde, N Ashish, G A Burns, and B N Nichols. Towards structured sharing of raw and derived neuroimaging data across existing resources. Neuroimage, 82:647–661, November 2013.
- D20
Camille Maumet, Tibor Auer, Alexander Bowring, Gang Chen, Samir Das, Guillaume Flandin, Satrajit Ghosh, Tristan Glatard, Krzysztof J Gorgolewski, Karl G Helmer, Mark Jenkinson, David B Keator, B Nolan Nichols, Jean-Baptiste Poline, Richard Reynolds, Vanessa Sochat, Jessica Turner, and Thomas E Nichols. Sharing brain mapping statistical results with the neuroimaging data model. Scientific Data, 3(1):1–15, December 2016.
- D21
Kyle Meyer, Michael Hanke, Yaroslav Halchenko, Benjamin Poldrack, and Adina Wagner. Datalad/datalad-container: 1.1.4. April 2021.
- D22
Clara A Moreau, Martineau Jean-Louis, Ross Blair, Christopher J Markiewicz, Jessica A Turner, Vince D Calhoun, Thomas E Nichols, and Cyril R Pernet. The genetics-BIDS extension: easing the search for genetic data associated with human brain imaging. Gigascience, October 2020.
- D23
Luc Moreau, Paul Groth, James Cheney, Timothy Lebo, and Simon Miles. The rationale of PROV. Web Semantics, 35:235–257, December 2015.
- D24
Guiomar Niso, Krzysztof J Gorgolewski, Elizabeth Bock, Teon L Brooks, Guillaume Flandin, Alexandre Gramfort, Richard N Henson, Mainak Jas, Vladimir Litvak, Jeremy T Moreau, Robert Oostenveld, Jan-Mathijs Schoffelen, Francois Tadel, Joseph Wexler, and Sylvain Baillet. MEG-BIDS: the brain imaging data structure extended to magnetoencephalography. Scientific Data, 5:180110, June 2018.
- D25
Brian A Nosek, Charles R Ebersole, Alexander C DeHaven, and David T Mellor. The preregistration revolution. Proceedings of the National Academy of Sciences, 2017(15):201708274, 2018.
- D26
Brian A Nosek and Daniel Lakens. Registered reports: a method to increase the credibility of published reports. Social Psychology, 45(3):137–141, 2014.
- D27
Brian A Nosek, Jeffrey R Spies, and Matt Motyl. Scientific utopia: II. restructuring incentives and practices to promote truth over publishability. Perspect. Psychol. Sci., 7(6):615–631, November 2012.
- D28
missing booktitle in Norgaard2019-su
- D29
Cyril R Pernet, Stefan Appelhoff, Krzysztof J Gorgolewski, Guillaume Flandin, Christophe Phillips, Arnaud Delorme, and Robert Oostenveld. EEG-BIDS: an extension to the brain imaging data structure for electroencephalography. Scientific data, 6(1):103, June 2019.
- D30
Russell A Poldrack, Chris I Baker, Joke Durnez, Krzysztof J Gorgolewski, Paul M Matthews, Marcus R Munafò, Thomas E Nichols, Jean Baptiste Poline, Edward Vul, and Tal Yarkoni. Scanning the horizon: towards transparent and reproducible neuroimaging research. Nat. Rev. Neurosci., 18(2):115–126, 2017.
- D31
Russell A Poldrack, Franklin Feingold, Michael J Frank, Padraig Gleeson, Gilles de Hollander, Quentin Jm Huys, Bradley C Love, Christopher J Markiewicz, Rosalyn Moran, Petra Ritter, Timothy T Rogers, Brandon M Turner, Tal Yarkoni, Ming Zhan, and Jonathan D Cohen. The importance of standards for sharing of computational models and data. Computational Brain & Behavior, 2(3-4):229–232, December 2019.
- D32
Russell A Poldrack, Grace Huckins, and Gael Varoquaux. Establishment of best practices for evidence for prediction: a review. JAMA Psychiatry, 77(5):534–540, May 2020.
- D33
Russell A Poldrack, Aniket Kittur, Donald Kalar, Eric Miller, Christian Seppa, Yolanda Gil, D Stott Parker, Fred W Sabb, and Robert M Bilder. The cognitive atlas: toward a knowledge foundation for cognitive neuroscience. Frontiers in Neuroinformatics, 5:17, September 2011.
- D34
Jean-Baptiste Poline, David N Kennedy, Friedrich T Sommer, Giorgio A Ascoli, David C Van Essen, Adam R Ferguson, Jeffrey S Grethe, Michael J Hawrylycz, Paul M Thompson, Russell A Poldrack, Satrajit S Ghosh, David B Keator, Thomas L Athey, Joshua T Vogelstein, Helen S Mayberg, and Maryann E Martone. Is neuroscience FAIR? a call for collaborative standardisation of neuroscience data. Neuroinformatics, January 2022.
- D35
K Robbins, D Truong, S Appelhoff, A Delorme, and S Makeig. Capturing the nature of events and event context using hierarchical event descriptors (HED). Neuroimage, pages 118766, November 2021.
- D36
K Robbins, Dung Truong, Alexander Jones, Ian Callanan, and Scott Makeig. Building FAIR functionality: annotating events in time series data using hierarchical event descriptors (HED). 2021.
- D37
D C Van Essen, Stephen M Smith, Deanna M Barch, Timothy E J Behrens, Essa Yacoub, Kamil Ugurbil, and WU-Minn HCP Consortium. The WU-Minn human connectome project: an overview. Neuroimage, 80:62–79, October 2013.
- D38
Matteo Visconti di Oleggio Castello, James E Dobson, Terry Sackett, Chandana Kodiweera, James V Haxby, Mathias Goncalves, Satrajit Ghosh, and Yaroslav O Halchenko. ReproNim/reproin 0.6.0. January 2020.
- D39
Adina S Wagner, Laura K Waite, Kyle Meyer, Marisa K Heckner, Tobias Kadelka, Niels Reuter, Alexander Q Waite, Benjamin Poldrack, Christopher J Markiewicz, Yaroslav O Halchenko, Peter Vavra, Pattarawat Chormai, Jean-Baptiste Poline, Lya K Paas, Peer Herholz, Lisa N Mochalski, Nevena Kraljevic, Lisa Wiersch, Alexandre Hutton, Dorian Pustina, Hamzah Hamid Baagil, Tristan Glatard, Sarah Oliveira, Giulia Ippoliti, Christian Mönch, Dorien Huijser, Surya T Togaru, Ariel Rokem, Rémi Gau, Judith Bomba, Małgorzata Wierzba, Stefan Appelhoff, Michael Joseph, and Michael Hanke. The DataLad handbook. 2021.
- D40
Mark D Wilkinson, Michel Dumontier, I Jsbrand Jan Aalbersberg, Gabrielle Appleton, Myles Axton, Arie Baak, Niklas Blomberg, Jan-Willem Boiten, Luiz Bonino da Silva Santos, Philip E Bourne, Jildau Bouwman, Anthony J Brookes, Tim Clark, Mercè Crosas, Ingrid Dillo, Olivier Dumon, Scott Edmunds, Chris T Evelo, Richard Finkers, Alejandra Gonzalez-Beltran, Alasdair J G Gray, Paul Groth, Carole Goble, Jeffrey S Grethe, Jaap Heringa, Peter A C 't Hoen, Rob Hooft, Tobias Kuhn, Ruben Kok, Joost Kok, Scott J Lusher, Maryann E Martone, Albert Mons, Abel L Packer, Bengt Persson, Philippe Rocca-Serra, Marco Roos, Rene van Schaik, Susanna-Assunta Sansone, Erik Schultes, Thierry Sengstag, Ted Slater, George Strawn, Morris A Swertz, Mark Thompson, Johan van der Lei, Erik van Mulligen, Jan Velterop, Andra Waagmeester, Peter Wittenburg, Katherine Wolstencroft, Jun Zhao, and Barend Mons. The FAIR guiding principles for scientific data management and stewardship. Scientific Data, 3:160018, March 2016.
- 1
Sources: Icons from the Noun Project: Structure by Adam Baihaqi from NounProject.com; Metadata by M. Oki Orlando; Data Management by ProSymbols; Logos: used with permission by the copyright holders.